We aim to understand the molecular pathways that control disease progression in bacterial pathogens. A major focus of the lab is to understand how regulatory non-coding RNAs control and co-ordinate expression of genes required for pathogensis and antibiotic resistance.
Regulatory RNAs and pathogenesis
Our model system is the human pathogen, enterohaemorrhagic E. coli O157 (EHEC), that causes disease ranging from gastroenteritis to life treating haemolytic uremic syndrome. The later is caused by release of Shiga toxins that are expressed from bacteriophage (bacterial viruses) that have inserted into the bacterial genome. We have recently identified large numbers of non-coding RNAs that are encoded in EHEC and are seeking to identify those that regulate virulence. We anticipate that by understanding how bacteria employ ncRNAs to control gene expression and respond to stress we will be able to design interventions that limit bacterial pathogenesis and dissemination.
Regulatory RNAs and antibiotic resistance
Non-coding RNAs play prominent roles in controlling the composition of the bacterial cell wall and membranes. A number of ncRNAs have been shown to control intrinsic antibiotic resistance in bacterial pathogens with ‘urgent’ or ‘serious’ levels of antimicrobial resistance. Non-coding RNAs contribute to antibiotic resistance in Methicillin-resistant Staphylococcus aureus (MRSA, designated a ‘serious’ threat) and we are using high-through put sequencing technologies, bioinformatics, and molecular biology to identify key ncRNAs that mediate antibiotic resistance in MRSA.
A little more detail…
Infectious diseases are among the leading causes of morbidity and mortality worldwide. Bacterial pathogens are highly adaptable and transfer of DNA between bacteria allows rapid acquisition of new traits/genes, particularly those under acute selective pressure like antibiotic resistance and pathogenesis in new hosts (i.e.: zoonoses). We are interested in how bacterial pathogens adapt with a focus on the mechanisms that pathogens use to control gene expression.
Enterohaemorrhagic E. coli
Enterohaemorhaggic E. coli (EHEC) is a pathogenic cousin of the harmless commensal E. coli that live in a healthy gastrointestinal tract. EHEC causes sporadic outbreaks of disease when it enters the food chain, often through contaminated meat or vegetables. Infection is associated with diarrheal disease, but can progress to potential fatal haemolytic uremic syndrome (kidney damage caused by Shiga toxins). Genome sequencing has demonstrated that the EHEC genome is 1.5 million base pairs larger than commensal E. coli and this extra DNA carries the genes that allow it to cause severe disease. We have identified 55 new regulatory non-coding RNAs in this 1.5Mb of pathogen-specific DNA.
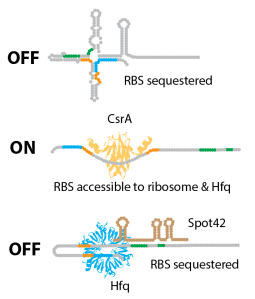
We are interested in how these regulatory RNAs turn virulence genes ‘on’ and ‘off’ and how they are co-ordinated during infection.
Multi-drug resistant Staphylococcus aureus
Staphylococcus aureus is one of the most adaptable and frequent human pathogens, causing disease at almost every site in the body. Multi-drug resistant S. aureus is increasingly common in community and healthcare settings and has been designated a “serious” threat by the Centre for Disease Control (CDC, USA). Approximately 17% of S. aureus blood infections in Australia are resistant to methicillin. These infections have a very high mortality rate (15-50%). The treatment of choice is the last line antibiotic vancomycin but up to 13% of MRSA isolates also display intermediate-resistance that leads to vancomycin treatment failure.
Intermediate-vancomycin resistance is associated with changes in the gene regulatory network. We are using the technical advances that we developed in EHEC to study non-coding RNA networks in MRSA, and understand how the gene regulatory network is different in intermediate-vancomycin resistant strains. Using these techniques we have found a new regulatory non-coding RNA pathway that is required for intermediate-vancomycin resistance.